安装包
- conda create -n seurat -c conda-forge r-seurat=4.1.1 -y
- conda activate seurat
- conda install -c conda-forge r-tidyverse -y
- conda install -c conda-forge r-irkernel -y
- Rscript -e “IRkernel::installspec(name=’seurat’, displayname=’r-seurat’)”
- conda install -c conda-forge r-biocmanager -y
- # conda install -c conda-forge r-sctransform -y
- # conda install -c conda-forge r-rocr -y
- conda install -c conda-forge r-modes -y
- # conda install -c conda-forge r-kernsmooth -y
- conda install -c conda-forge r-fields -y
- conda install -c conda-forge r-devtools -y
- conda install -c conda-forge r-clustree -y
- BiocManager::install(“glmGamPoi”)
- ~/dev/xray/xray -c ~/etc/xui2.json &
- wget -e “https_proxy=http://127.0.0.1:20809“ https://github.com/chris-mcginnis-ucsf/DoubletFinder/archive/refs/heads/master.zip -O DoubletFinder-master.zip
- devtools::install_local(‘DoubletFinder-master.zip’)
读取数据
从UMI矩阵
1
2
3
4
5
6
7
8
9
10
11
| sce <- read.table(gzfile("GSM4203181_data.matrix.txt.gz"),
header = T, row.names = 1)
sce <- Seurat::CreateSeuratObject(sce, project = 'GSM4203181',
min.cells = 3, min.features = 200)
sce[["percent.mt"]] <- Seurat::PercentageFeatureSet(sce, pattern = "^MT-")
sce[["percent.ERCC"]] <- Seurat::PercentageFeatureSet(sce, pattern = "^ERCC-")
tmp <- rownames(sce@meta.data)
sce[["orig.ident"]] <- paste0('sample', substr(tmp,18,nchar(tmp)))
table(sce[["orig.ident"]])
sce@meta.data
saveRDS(sce, 'GSM4203181.rds')
|
cellranger输出
1
2
3
4
5
| sce <- Seurat::Read10X('filtered_feature_bc_matrix')
sce <- Seurat::CreateSeuratObject(sce, project = 'SRX6887740',
min.cells = 3, min.features = 200)
sce[["percent.mt"]] <- Seurat::PercentageFeatureSet(sce, pattern = "^MT-")
sce[["percent.ERCC"]] <- Seurat::PercentageFeatureSet(sce, pattern = "^ERCC-")
|
初步QC
1
2
3
4
5
6
| options(repr.plot.width = 12, repr.plot.height = 6)
Seurat::VlnPlot(sce, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
plot1 <- Seurat::FeatureScatter(sce, feature1 = "nCount_RNA", feature2 = "percent.mt")
plot2 <- Seurat::FeatureScatter(sce, feature1 = "nCount_RNA", feature2 = "nFeature_RNA")
plot1 + plot2
sce <- subset(sce, nFeature_RNA > 2500 & nFeature_RNA < 7500 & nCount_RNA > 2500 & nCount_RNA < 50000 & percent.mt < 7.5)
|
计算细胞周期评分
1
2
3
4
5
6
7
| sce <- Seurat::NormalizeData(sce)
g2m_genes <- Seurat::CaseMatch(search=Seurat::cc.genes$g2m.genes,
match=rownames(sce))
s_genes <- Seurat::CaseMatch(search=Seurat::cc.genes$s.genes,
match=rownames(sce))
sce <- Seurat::CellCycleScoring(sce, g2m.features=g2m_genes, s.features=s_genes)
sce$CC.Difference <- sce$S.Score - sce$G2M.Score
|
1
2
3
| sce <- Seurat::SCTransform(sce, vst.flavor = "v2",
vars.to.regress = c("CC.Difference", "percent.mt"),
verbose = F)
|
查看分群走向
降维
1
2
3
4
5
6
| sce <- Seurat::RunPCA(sce, assay="SCT", verbose = FALSE)
Seurat::ElbowPlot(sce, ndims = 50)
sce <- Seurat::RunUMAP(sce, reduction = "pca",
dims = 1:30, verbose = FALSE)
sce <- Seurat::FindNeighbors(sce, reduction = "pca",
dims = 1:30, verbose = FALSE)
|
分群
1
2
3
4
5
6
7
| sce <- Seurat::FindClusters(
object = sce,
resolution = c(seq(.1,1.6,.2))
)
options(repr.plot.width = 12, repr.plot.height = 12)
require(clustree)
clustree::clustree(sce@meta.data, prefix = "SCT_snn_res.")
|
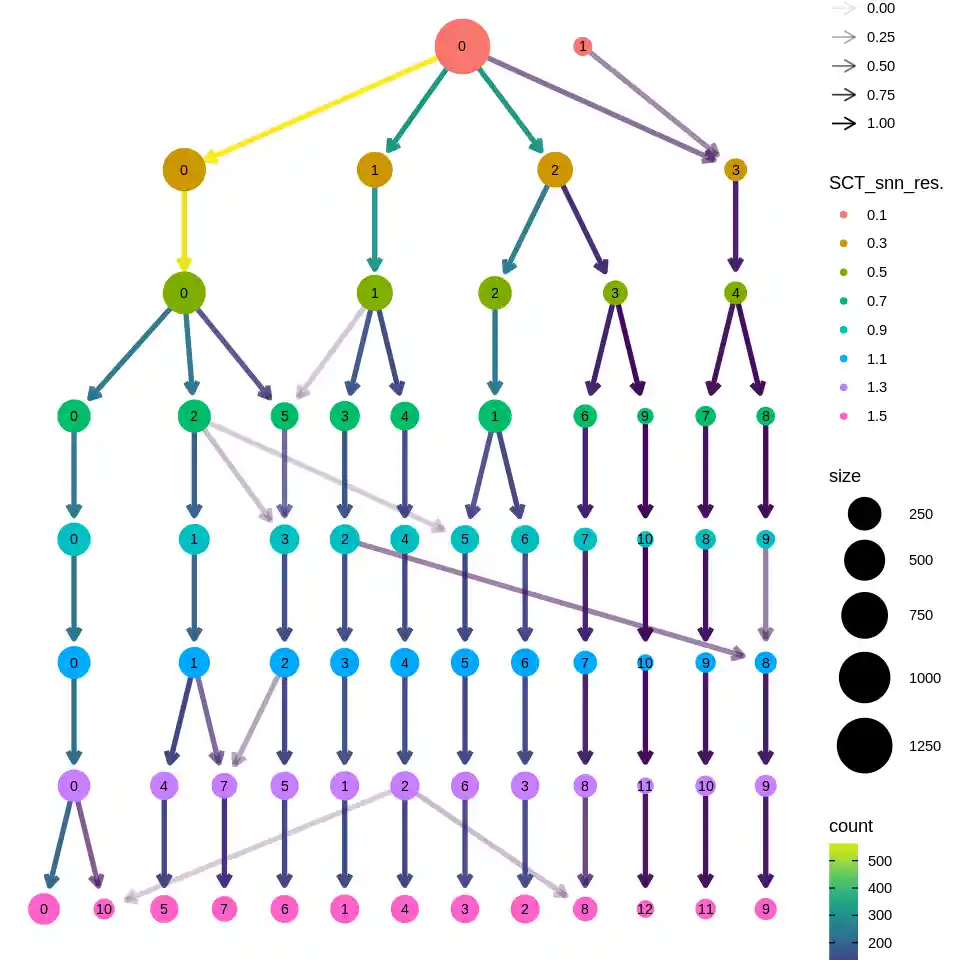
这图还挺好看的
使用DoubletFinder去除doublet
pK Identification
1
2
3
| sweep.res <- DoubletFinder::paramSweep_v3(sce, PCs = 1:10, sct = T, num.cores=8)
sweep.stats <- DoubletFinder::summarizeSweep(sweep.res, GT = FALSE)
bcmvn <- DoubletFinder::find.pK(sweep.stats)
|
Run DoubletFinder
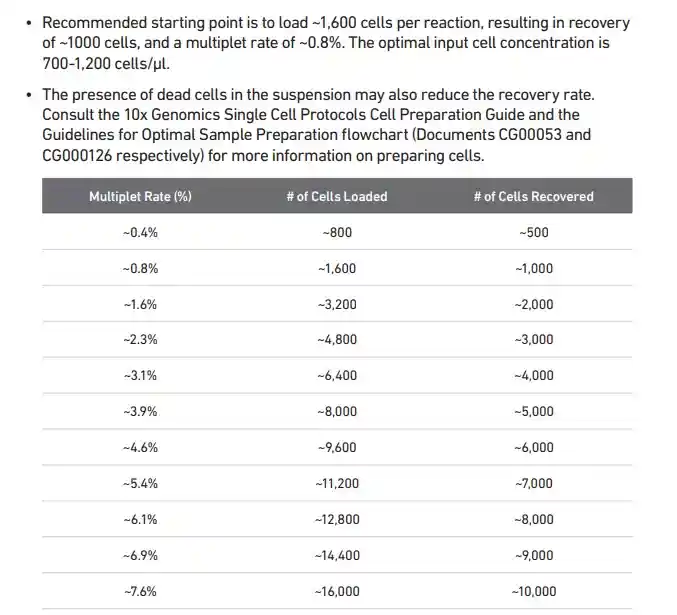
10X genomics单细胞平台的dulblets比率
1
2
3
4
5
6
7
8
9
|
nExp_poi <- round(0.016*nrow(sce@meta.data))
pK_bcmvn <- as.numeric(as.character(bcmvn$pK[which.max(bcmvn$BCmetric)]))
sce <- DoubletFinder::doubletFinder_v3(sce, PCs = 1:10,
pN = 0.25, pK = pK_bcmvn,
nExp = nExp_poi, reuse.pANN = FALSE,
sct = T)
sce@meta.data
DimPlot(sce, reduction = "umap", group.by = c("DF.classifications_0.25_0.2_20"))
|
1
2
| sce <- subset(sce, DF.classifications_0.25_0.2_20 != 'Doublet')
saveRDS(sce, 'SRX6887740.rds')
|

